This blog post was co-authored with Bianca Furtuna and Prasanna Muralidharan.
Introduction
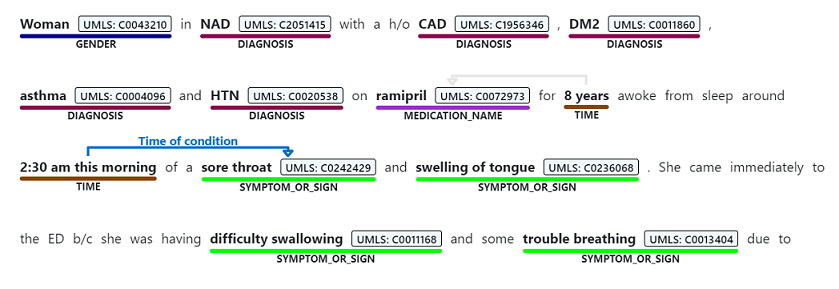
Information extraction is the process of extracting entities, relations, assertions, topics, and additional information from textual data. For example, we may want to extract medical information from doctors’ clinical notes (See figure 1) and later correlate that with the patient health trajectory. Similarly, we may want to extract topics out of financial reports written by market analysts for information retrieval. In Microsoft’s Commercial Software Engineering team (CSE), we often collaborate with strategic customers on information extraction problems such as Named Entity Recognition (NER). Each new project encompasses specific ways of gathering the raw data, multiple models to experiment with, and different strategies for evaluating the result. In addition, different customers will have their data stored differently and are likely to have different requirements on how these systems should be tested and evaluated.
The approach we take for each project follows best practices for building and deploying Machine Learning (ML) applications encompassing hypotheses, experiments, data, code, and models of high quality that can be reliably managed and reproduced. These best practices ensure that a structured and well-defined experimentation and evaluation flow is set up at the beginning of the project. Such setup enables a clear definition of how performance will be measured, and how all models are compared in a consistent manner. In addition, it allows for error analysis for each candidate model or models and can support models in any language.
In this blog post we cover the process, requirements, and the design of such an evaluation framework for Information Extraction and specifically Named Entity Recognition.
Why build an evaluation framework?
Evaluating ML models is hard! and evaluation logic can become very complex. In real world cases, the implementation of the evaluation logic should be agreed upon by various stakeholders to make sure the model is optimized on the right business and technical metrics.
To overcome these challenges, we propose an evaluation framework that is modular and performs robust and consistent evaluations. The framework has the following characteristics:
- Allows for experimentation with different datasets and models.
- Facilitates collaboration of multiple team members as it ensures a consistent evaluation flow which enables team members to work together on developing the model.
- Easy integration into MLOps frameworks for continuous evaluation in a production setting.
- Providing the capability to perform exhaustive tests of evaluation logic.
Design
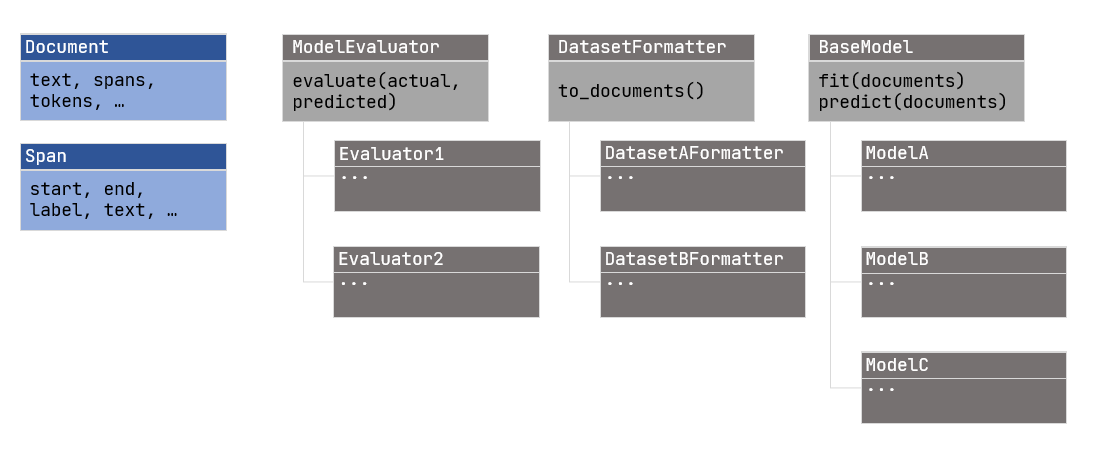
The framework is provided as a python package with four main parts:
- An internal representation (of documents, spans, and tokens)
- Abstract class for formatting datasets (DatasetFormatter)
- Abstract class for models (BaseModel)
- Abstract class for evaluation logic (ModelEvaluator)
The different classes are shown in Figure 2 – Class Diagram.
Let’s go deeper into what each part looks like:
Data Objects
The framework contains data objects which are passed between the different modules. These classes represent standard NLP objects like raw text, spans, and tokens. Objects can be serialized and translated to different representations.
The main data object is the Document class. The Document class has the following fields:
- Document id
- Spans: holding the start and end of entities or phrases
- Text: The raw text of this document
- Tokens: A tokenized representation of the raw text, using a predefined tokenizer
- Metadata: Additional metadata on the document
The Document objects is resembling objects used by other frameworks, such as spaCy and Prodigy:
{
"spans": [{
"start": 11,
"end": 15,
"label": "PERSON",
"token_start": 3,
"token_end": 3,
"text": "Lisa"
}
],
"tokens": ["My", "name", "is", "Lisa"],
"meta": {
"id": 0
},
"text": "My name is Lisa"
}
For Named Entity Recognition, the Document and Span objects can be translated from/into BIO/IOB and BILUO/BIOES, allowing easy integration into models which expect such input or datasets in this structure.
Dataset Formatter
The formatter abstraction is used to translate any given input data into a unified data representation. Its implementation should include the loading of the original dataset from a file or a stream, and the translation logic into an Iterable of Document objects. Such abstraction permits the evaluation of models on multiple types of datasets, while still maintaining one evaluation flow. Here’s the DatasetFormatter code:
class DatasetFormatter(ABC):
@abstractmethod
def to_documents(self) -> Iterable[Document]:
"""
Translate a dataset structure into an iterable of documents,
to be used by models and for evaluation
"""
pass
Base Model
The model abstraction allows a user to experiment with multiple types of models. For example, we can wrap a CRF model, a spaCy model and a PyTorch model and compare the three given the same dataset and the same evaluator:
class BaseModel(ABC):
def __init__(self):
pass
@abstractmethod
def fit(self, documents: Iterable[Document]):
pass
@abstractmethod
def predict(self, documents: Iterable[Document]):
pass
Model Evaluator
The ModelEvaluator abstraction allows a user to implement different evaluation strategies, while re-using previously implemented dataset connectors or models. We can further use it to evaluate already existing predictions from a file generated previously or outside this framework.
class ModelEvaluator(ABC):
"""
Abstract class to hold logic for evaluation functions
"""
@abstractmethod
def evaluate(
self, annotations: Iterable[Document], predictions: Iterable[Document]
) -> Dict:
"""
Evaluate a model on a corpus of documents, each containing annotated and predicted spans,
and return the overall model metrics
:param annotations: List of documents with annotated spans
:param predictions: List of documents with predicted spans
:return: metrics
"""
pass
Example use case: Detecting private entities in text
Detecting names, organizations and locations is a common problem in NLP. Using this evaluation framework, we can evaluate different NER models on different datasets, using a predefined evaluation strategy.
Datasets
In case we have multiple datasets in different formats, we can create different formatters. One common format for NER is the one used in the CoNLL-2003 shared task. Another common format is the brat standoff format. Different annotation tools like Prodigy or Doccano have their own JSONL output format.
Let’s assume we want to train a model on a brat standoff formatted dataset, and then fine tune it on manually labeled data. Using the `DatasetFormatter` object, we would create two classes: BratFormatter and DoccanoFormatter. The logic for transforming each type of data into the unified representation of List[Document] will be written in the to_documents function in each class. In addition, we use spaCy to tokenize the input dataset.
Here’s a naïve implementation of the BratFormatter:
from pathlib import Path
from nlp_eval import Span, Document
from nlp_eval.formatting import DatasetFormatter
class BratFormatter(DatasetFormatter):
def __init__(self, files_path: Path):
"""
Translator between the brat standoff format (https://brat.nlplab.org/standoff.html)
to the internal representation of this package
:param files_path Path containing txt and ann files.
"""
super().__init__()
self.files_path = files_path
def to_documents(self):
for txt_path in Path(self.files_path).glob("*.txt"):
document_id = txt_path.stem
annotation_file_path = Path(self.files_path, f"{document_id}.ann").resolve()
with open(str(annotation_file_path), encoding="utf-8-sig") as f_ann:
ann = f_ann.readlines()
with open(str(txt_path), encoding="utf-8-sig") as f_txt:
text = f_txt.read().replace('\n', ' ')
spans = []
for line in ann:
annotation = line.split()
spans.append(
Span(
label=annotation[1],
start=int(annotation[2]),
end=int(annotation[3]) - 1,
text=" ".join(annotation[4:]),
)
)
yield Document(text=text, spans=spans, document_id=document_id, tokens=self.nlp(text))
Now that we have our two datasets ready for modeling, let’s discuss the different models we’d like to experiment with.
Models
Let’s assume we want to experiment with three models – a simple CRF model (using the sklearn-crfsuite package), a spaCy model and finally a transformer-based model using packages like Flair or transformers. We can use the BaseModel abstract class to form three new classes, one for each model type. This would allow us to experiment with the different models using the exact same flow, and would therefore create a consistent API for all models. While these model wrappers are an overhead, they help assure that we are comparing apples to apples, and would help if we decide to change our production model in our pipeline from one to the other, as they all have identical APIs.
The two abstract methods in BaseModel are fit and predict, which come from the popular scikit-learn package. We would use each method to translate the unified Document representation into the expected input of the specific model and would also translate the output of the model into the same representation so that we could use a unified evaluation for all models.
Here’s a small example on how to adapt a spaCy NER model into a BaseModel:
class SpacyNERModel(BaseModel):
def __init__(self, nlp=None):
self.nlp = nlp
super().__init__()
def fit(self, documents: Iterable[Document]):
# spaCy simple training style, taken from https://spacy.io/usage/training#training-simple-style
train_data = SpacyNERModel._documents_to_spacy_train_data(documents)
if not self.nlp:
self.nlp = spacy.blank("en")
optimizer = self.nlp.begin_training()
for i in range(20):
random.shuffle(train_data)
for text, annotations in train_data:
self.nlp.update([text], [annotations], sgd=optimizer)
self.nlp.to_disk("/model")
def predict(self, documents: Iterable[Document]):
doc_tuples = [(d.text, d) for d in documents]
for doc, original_doc in self.nlp.pipe(doc_tuples, as_tuples=True):
predicted_spans = [Span.from_spacy_span(ent) for ent in doc.ents]
spacy_doc = Document(text=original_doc.text, tokens=doc, spans=predicted_spans)
yield spacy_doc
@staticmethod
def _documents_to_spacy_train_data(documents: Iterable[Document]):
training_data = []
for document in documents:
document.handle_overlapping_spans()
training_data.append((document.text, SpacyNERModel._spans_to_spacy_spans(document.spans)))
return training_data
@staticmethod
def _spans_to_spacy_spans(spans: List[Span]):
return {"entities": [(span.start, span.end, span.label) for span in spans]}
On the fit function, we translate the documents into the requested input by spaCy. Then we train a spaCy model and save it. On the predict function, we run batch prediction on the test documents, and translate the output into a list of Document objects.
Evaluation
Lastly, for evaluation, we could use the ModelEvaluator abstract class to implement the evaluation we require. Since in most cases we would only have one evaluation strategy, we can consider either using the evaluator object directly (making it non-abstract) or create a class which implements it.
In this scenario, let’s use the seqeval package to calculate different NER metrics.
class NERSimpleEvaluator(ModelEvaluator):
"""
Contains the evaluator functions for Named Entity Recognition (NER) using seqeval framework
"""
def evaluate(self, annotations: Iterable[Document], predictions: Iterable[Document], schema=BILOU):
"""
Evaluate a model on a corpus of documents, each containing annotated and predicted spans
"""
annot_list = []
pred_list = []
for annot, pred in zip(annotations, predictions):
annot_list.append(annot.get_biluo())
pred_list.append(pred.get_biluo())
return classification_report(
annot_list, pred_list, mode="strict", scheme=schema
)
Experiment flow
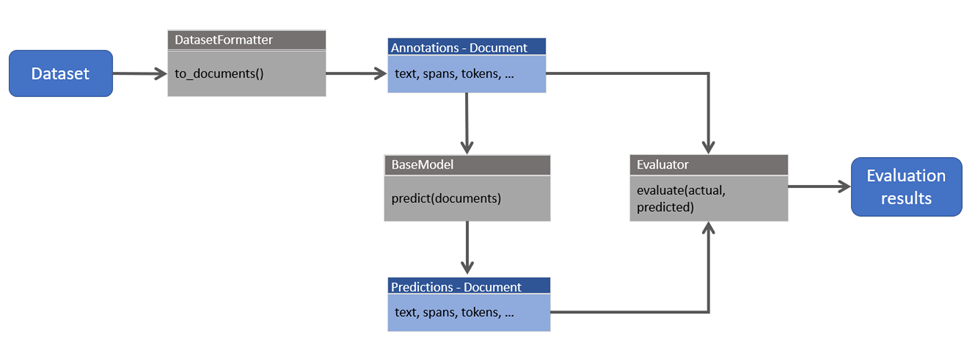
Now that we have the different building blocks defined, let’s look at an example end-to-end flow (summarized in the sequence diagram below):
Here is an example flow in code, using a formatter for loading the CONLL2003 dataset and the SpacyNERModel:
1. Read dataset into a list of `Document` objects:
formatter = CONLL2003Formatter() formatter.download() doc_iter = formatter.to_documents(fold="eng.testb") # run flow only on a subset of the examples documents = list(doc_iter)[:10]
2. Optionally save the dataset to JSONL format
Document.save_dataset(
documents=documents, output_format="jsonl", output_file="testa.jsonl"
)
3. Load model for prediction
model = SpacyNERModel(nlp='mymodel') predicted_docs = model.predict(documents)
4. Evaluate results
evaluator = NEREvaluator() results = evaluator.evaluate(annotations=documents, predictions=predicted_docs) evaluator.print_results_dict()
Another example flow is when we already have predictions stored in a file, and we use the Evaluator to compare stored annotations with stored predictions.
Advantages of using the evaluation framework
Reproducibility
When leveraging a structured evaluation pipeline, it is straightforward to achieve full reproducibility of experiment runs. For this purpose we used MLFlow to track dataset identifiers, hyper parameters and outputted metrics for every experiment run, and collected a comparable set of results for experiments with different model types.
Operationalization
The proposed evaluation framework can be installed as a Python package. As such, it can be used in production environments to evaluate models or installed by individual team members for individual experimentation. Since the models in the package all inherit from the BaseModel class, they are easily interchangeable, so one could replace the model in the production pipeline with another, without having to create adapters for the new model.
Testing
By creating evaluators, models and datasets as defined objects, unit testing and other forms of testing becomes easier. For example, we could create a unit test for the to_documents function on DatasetFormatter, verifying it reads the data correctly. We could do simple tests to `fit` and `predict` on real or mock models, and we can thoroughly test our evaluation strategy, to verify we’re not getting wrong results due to bugs in evaluation.
For example, here’s one unit-test for validating that metrics (precision, recall and f1 loaded from the metrics object) are calculated correctly:
@pytest.mark.parametrize(
"tp, fp, fn, tn, e_prec, e_rec, e_f1",
[ # simple example
(4, 1, 4, 3, 0.8, 0.5, 8 / 13),
# zero edge case
(0, 0, 0, 0, 0, 0, 0),
# some zero counts
(0, 0, 4, 23, 0, 0, 0),
],
)
def test_calculate_metrics_returns_correct_values(tp, fp, fn, tn, e_prec, e_rec, e_f1):
metrics = ModelMetrics.calculate_metrics(tp, fp, fn, tn)
assert metrics.precision == e_prec
assert metrics.recall == e_rec
assert metrics.f_1 == e_f1
Summary
Experimenting with ML models is always a challenge. Data scientists often use models from different frameworks, write custom logic and have different assumptions when building the experimentation pipelines. Often, the data scientist owns the evaluation flow from data collection through modeling to results, and different team members working on the same problem might evaluate their model in a different way. Lastly, potential bugs in the flow or specifically in the evaluation code could cause the experiment results to be wrong, which might lead to wrong conclusions or time wasted on experimentation. Mainly for those reasons, we propose a more structured way of experimenting with and evaluating models.
Creating an evaluation framework for your ML project is a great step towards rigorous experimentation and operationalization. The proposed framework introduces structure into the experiment flow, and creates consistency for using different datasets, models, or evaluators. The main limitation however is the overhead in translating data and model APIs between the original format to the framework’s format.
Icon made by Becris, Icongeek26, Linector and Freepik from www.flaticon.com.